Project A1
Cross-talk of Aspergillus fumigatus with neutrophilic granulocytes
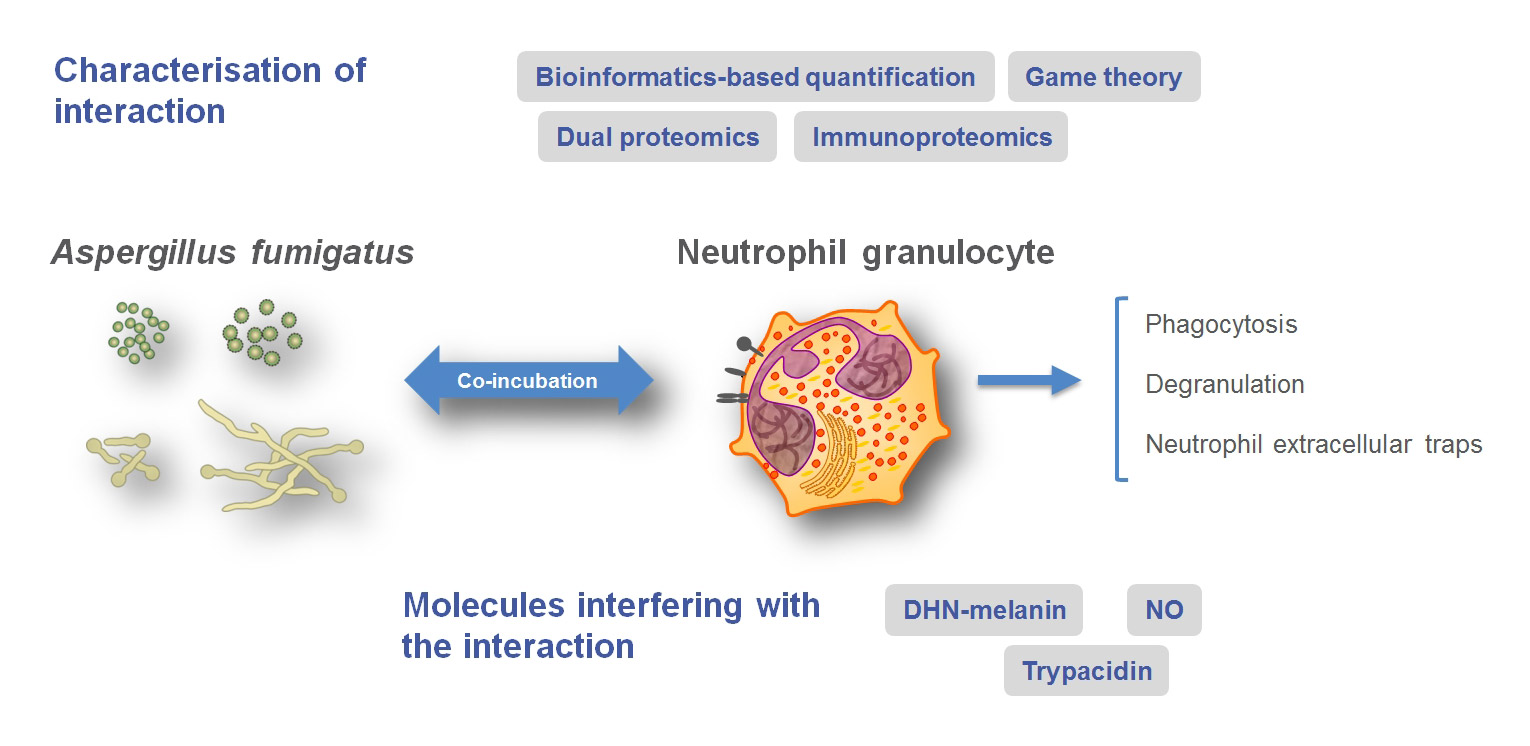
Neutrophils are the most important effector cells in the immune response against Aspergillus fumigatus. In project A1, we aim at understanding the mechanisms how neutrophils control A. fumigatus infections.
In the first funding period we developed novel methods for the bioinformatics-based analysis of live cell imaging. This allowed automated quantification of phagocytosis and the identification of fungal molecules inhibiting phagocytosis. Furthermore, quantitative analysis of the dual proteome of A. fumigatus and neutrophils by LC-MS was established. This led to the discovery of novel human proteins involved in the interactions with this fungus.
In the second funding period our research will focus on the identification of novel effector mechanisms of neutrophils against A. fumigatus, building on our discoveries from the first funding period. We will also characterize immune cell receptors potentially involved in the cross-talk of neutrophils with A. fumigatus. For this, we will use immune cells from knockout mice, cell lines, monoclonal antibodies that block specific receptors and mouse infection models for invasive aspergillosis. Finally, we will analyze patient groups for single nucleotide polymorphisms (SNPs) in the proteins that interact with A. fumigatus as basis for translational approaches.

Prof. Dr. Axel Brakhage
Project A1 / Project A6 / Project Z1
Institute of Microbiology
Friedrich Schiller University Jena
Department of Molecular and Applied Microbiology HKI
Leibniz Institute for Natural Product Research and Infection Biology - Hans-Knöll-Institute

Prof. Marie Lilienfeld-Toal
Institute for Diversity Medicine, Faculty of Medicine
Ruhr University Bochum
| Author | Year | Title | Journal | Links |
|---|---|---|---|---|
| Müller R, König A, Groth S, Zarnowski R, Visser C, Handrianz T, Maufrais C, Krüger T, Himmel M, Lee S, Priest EL, Yildirim D, Richardson JP, Blango MG, Bougnoux ME, Kniemeyer O, d'Enfert C, Brakhage AA, Andes DR, Trümper V, Nehls C, Kasper L, Mogavero S, Gutsmann T, Naglik JR, Allert S, Hube B | 2024 | Secretion of the fungal toxin candidalysin is dependent on conserved precursor peptide sequences. | Nat Microbiol. (3):669-683. | PubMed |
| Goldmann M, Schmidt F, Cseresnyés Z, Orasch T, Jahreis S, Hartung S, Figge MT, von Lilienfeld-Toal M, Heinekamp T, Brakhage AA | 2023 | The lipid raft-associated protein stomatin is required for accumulation of dectin-1 in the phagosomal membrane and for full activity of macrophages against Aspergillus fumigatus. | mSphere 8(1) :e0052322 | PubMed |
| Orasch T, Gangapurwala G, Vollrath A, González K, Alex J, De San Luis A, Weber C, Hoeppener S, Cseresnyés Z, Figge MT, Guerrero-Sanchez C, Schubert US, Brakhage AA | 2023 | Polymer-based particles against pathogenic fungi: A non-uptake delivery of compounds. | Biomater Adv 146, 213300 | PubMed |
| Jia LJ, Rafiq M, Radosa L, Hortschansky P, Cunha C, Cseresnyés Z, Krüger T, Schmidt F, Heinekamp T, Straßburger M, Löffler B, Doenst T, Lacerda JF, Campos A, Figge MT, Carvalho A, Kniemeyer O, Brakhage AA | 2023 | Aspergillus fumigatus hijacks human p11 to redirect fungal-containing phagosomes to non-degradative pathway. | Cell Host & Microbe: 31,3 | Cell Host & Microbe |
| Kelani AA, Bruch A, Rivieccio F, Visser C, Krueger T, Weaver D, Pan X, Schaeuble S, Panagiotou G, Kniemeyer O, Bromley MJ, Bowyer P, Barber AE, Brakhage AA, Blango MG. | 2023 | Disruption of the Aspergillus fumigatus RNA interference machinery alters the conidial transcriptome. | RNA [Epub ahead of print] | PubMed |
| Sala A, Ardizzoni A, Spaggiari L, Vaidya N, van der Schaaf J, Rizzato C, Cermelli C, Mogavero S, Krüger T, Himmel M, Kniemeyer O, Brakhage AA, King BL, Lupetti A, Comar M, de Seta F, Tavanti A, Blasi E, Wheeler RT, Pericolini E. A | 2023 | New Phenotype in Candida-Epithelial Cell Interaction Distinguishes Colonization- versus Vulvovaginal Candidiasis-Associated Strains. | mBio 3: e0010723 | PubMed |
| Brioli A, Gengenbach L, Mancuso K, Binder M, Ernst T, Heidel FH, Stauch T, Zamagni E, Hilgendorf I, Hochhaus A, Engelhardt M, von Lilienfeld-Toal M. | 2023 | Pomalidomide combinations are a safe and effective option after daratumumab failure. | J Cancer Res Clin Oncol. [2]13 | PubMed |
| Giesen N, Busch E, Schalk E, Beutel G, Rüthrich MM, Hentrich M, Hertenstein B, Hirsch HH, Karthaus M, Khodamoradi Y, Koehler P, Krüger W, Koldehoff M, Krause R, Mellinghoff SC, Penack O, Sandherr M, Seggewiss-Bernhardt R, Spiekermann K, Sprute R, Stemler J, Weissinger F, Wörmann B, Wolf HH, Cornely OA, Rieger CT, von Lilienfeld-Toal M | 2023 | AGIHO guideline on evidence-based management of COVID-19 in cancer patients: 2022 update on vaccination, pharmacological prophylaxis and therapy in light of the omicron variants. | Eur J Cancer. 181: 102-118 | PubMed |
| Krespach MKC, Stroe MC, Netzker T, Rosin M, Zehner LM, Komor AJ, Beilmann JM, Krüger T, Scherlach K, Kniemeyer O, Schroeckh V, Hertweck C, Brakhage AA | 2023 | Streptomyces polyketides mediate bacteria–fungi interactions across soil environments. | Nature Microbiology | Nat Microb |
| Garloff V, Krüger T, Brakhage A, Rubio I. | 2023 | Control of TurboID-dependent biotinylation intensity in proximity ligation screens. J Proteomics. | J Proteomics 15;279:104886 | PubMed |
| Mirhakkak MH, Chen X, Ni Y, Heinekamp T, Sae-Ong T, Xu LL, Kurzai O, Barber AE, Brakhage AA, Boutin S, Schäuble S, Panagiotou G | 2023 | Genome-scale metabolic modeling of Aspergillus fumigatus strains reveals growth dependencies on the lung microbiome. | Nat Commun. 14(1):4369 | PubMed |
| Wein P, Dornblut K, Herkersdorf S, Krüger T, Molloy EM, Brakhage AA, Hoffmeister D, Hertweck C | 2023 | Bacterial secretion systems contribute to rapid tissue decay in button mushroom soft rot disease. | mBio: e0078723 | PubMed |
| Garloff V, Krüger T, Brakhage A, Rubio I. | 2023 | Control of TurboID-dependent biotinylation intensity in proximity ligation screens. | J Proteomics 279:104886 | PubMed |
| Sala A, Ardizzoni A, Spaggiari L, Vaidya N, van der Schaaf J, Rizzato C, Cermelli C, Mogavero S, Krüger T, Himmel M, Kniemeyer O, Brakhage AA, King BL, Lupetti A, Comar M, de Seta F, Tavanti A, Blasi E, Wheeler RT, Pericolini E | 2023 | A new phenotype in Candida-epithelial cell interaction distinguishes colonization-versus culvovaginal candidiasis-associated strains. | mBio 14(2), e0010723 | PubMed |
| Ramírez-Zavala B, Betsova D, Schwanfelder S, Krüger I, Mottola A, Krüger T, Kniemeyer O, Brakhage AA, Morschhäuser J. | 2023 | Multiple phosphorylation sites regulate the activity of the repressor Mig1 in Candida albicans. | mSphere: 8 (6) | PubMed |
| Schruefer S, Pschibul A, Wong SSW, Sae-Ong T, Wolf T, Schäuble S, Panagiotou G, Brakhage AA, Aimanianda V, Kniemeyer O, Ebel F | 2023 | Distinct transcriptional responses to fludioxonil in Aspergillus fumigatus and its ΔtcsC and Δskn7 mutants reveal a crucial role for Skn7 in the cell wall reorganizations triggered by this antifungal. | BMC Genomics. 14;24(1):684 | PubMed |
| Halder LD, Babych S, Palme DI, Mansouri-Ghahnavieh E, Ivanov L, Ashonibare V, Langenhorst D, Prusty B, Rambach G, Wich M, Trinks N, Blango MG, Kornitzer D, Terpitz U, Speth C, Jungnickel B, Beyersdorf N, Zipfel PF, Brakhage AA, Skerka C | 2022 | Candida albicans induces cross-kingdom miRNA trafficking in human monocytes to promote fungal growth. | mBio 13(1): e0356321 | mBio |
| Rafiq M, Rivieccio F, Zimmermann AK, Visser C, Bruch A, Krüger T, González Rojas K, Kniemeyer O, Blango MG, Brakhage AA | 2022 | PLB-985 neutrophil-like cells as a model to study Aspergillus fumigatus pathogenesis. | mSphere e0094021 | PubMed |
| Hoang TNM, Cseresnyés Z, Hartung S, Blickensdorf M, Saffer C, Rennert K, Mosig AS, von Lilienfeld-Toal M, Figge MT | 2022 | Invasive aspergillosis-on-chip: A quantitative treatment study of human Aspergillus fumigatus infection | Biomaterials 283: 121420 | PubMed |
| Brandt P, Gerwien F, Wagner L, Krüger T, Ramirez-Zavala B, Mirhakkak MH, Schäuble S, Kniemeyer O, Panagiotou G, Brakhage AA, Morschhäuser J, Vylkova S | 2022 | Candida albicans SR-Like Protein Kinases Regulate Different Cellular Processes: Sky1 Is Involved in Control of Ion Homeostasis, While Sky2 Is Important for Dipeptide Utilization. | Frontiers Cellular Infection Microbiology doi:0.3389/fcimb.2022.850531 | Front. Cell. Infect. Microbiol. |
| Fendler A, de Vries EGE, GeurtsvanKessel CH, Haanen JB, Wörmann B, Turajlic S, von Lilienfeld-Toal M | 2022 | COVID-19 vaccines in patients with cancer: immunogenicity, efficacy and safety. | Nat Rev Clin Oncol , 1-17. | PubMed |
| Goldman JD, Gonzalez MA, Rüthrich MM, Sharon E, von Lilienfeld-Toal M. | 2022 | COVID-19 and Cancer: Special Considerations for Patients Receiving Immunotherapy and Immunosuppressive Cancer Therapies. | Am Soc Clin Oncol Educ Book doi: 10.1200/EDBK 359656. | PubMed |
| Niemiec MJ, Kapitan M, Himmel M, Döll K, Krüger T, Köllner TG, Auge I, Kage F, Alteri CJ, Mobley HLT, Monsen T, Linde S, Nietzsche S, Kniemeyer O, Brakhage AA, Jacobsen ID | 2022 | Augmented Enterocyte Damage During Candida albicans and Proteus mirabilis Coinfection. | Front Cell Infect Microbiol. 12:866416 | PubMed |
| Balkenhol J, Bencurova E, Gupta SK, Schmidt H, Heinekamp T, Brakhage AA, Pottikkadavath A, Dandekar T | 2022 | Prediction and validation of host-pathogen interactions by a versatile inference approach using Aspergillus fumigatus as a case study. | Computational and Structural Biotechnology Journal 20: 4225-4237 | PubMed |
| Tappe B, Lauruschkat CD, Strobel L, Pantaleón García J, Kurzai O, Rebhan S, Kraus S, Pfeuffer-Jovic E, Bussemer L, Possler L, Held M, Hünniger K, Kniemeyer O, Schäuble S, Brakhage AA, Panagiotou G, White PL, Einsele H, Löffler J, Wurster S. | 2022 | COVID-19 patients share common, corticosteroid-independent features of impaired host immunity to pathogenic molds. | Front Immunol. 13: 954985 | PubMed |
| Böttcher B, Driesch D, Krüger T, Garbe E, Gerwien F, Kniemeyer O, Brakhage AA, Vylkova S | 2022 | Impaired amino acid uptake leads to global metabolic imbalance of Candida albicans biofilms. | NPJ Biofilms Microbiomes. 8(1): 78 | PubMed |
| Montaño DE, Hartung S, Wich M, Ali R, Jungnickel B, von Lilienfeld-Toal M, Voigt K | 2022 | The TLR-NF-kB axis contributes to the monocytic inflammatory response against a virulent strain of Lichtheimia corymbifera, a causative agent of invasive mucormycosis. | Front Immunol. 13: 882921 | PubMed |
| de Assis LJ, Silva LP, Bayram O, Dowling P, Kniemeyer O, Krüger T, Brakhage AA, Chen Y, Dong L, Tan K, Wong KH, Ries LNA, Goldman GH | 2021 | Carbon catabolite repression in filamentous fungi is regulated by phosphorylation of the transcription factor CreA. | mBio 12: e03146-20 | PubMed |
| Yu Y, Wolf AK, Thusek S, Heinekamp T, Bromley M, Krappmann S, Terpitz U, Voigt K, Brakhage AA, Beilhack A | 2021 | Direct visualization of fungal burden in filamentous fungus-infected silkworms. | J Fungi (Basel) 7: 136 | PubMed |
| Strømland Ø, Kallio JP, Pschibul A, Skoge RH, Harðardóttir HM, Sverkeli LJ, Heinekamp T, Kniemeyer O, Migaud M, Makarov MV, Gossmann TI, Brakhage AA, Ziegler M | 2021 | Discovery of fungal surface NADases predominantly present in pathogenic species. | Nat Commun 12: 1631 | PubMed |
| Lauruschkat CD, Page L, White PL, Etter S, Davies HE, Duckers J, Ebel F, Schnack E, Backx M, Dragan M, Schlegel N, Kniemeyer O, Brakhage AA, Einsele H, Loeffler J, Wurster S | 2021 | Development of a simple and robust whole blood assay with dual co-stimulation to quantify the release of T-cellular signature cytokines in response to Aspergillus fumigatus antigens. | J Fungi 7: 462 | MDPI |
| Page L, Wallstabe J, Lother J, Bauser M, Kniemeyer O, Strobel L, Voltersen V, Teutschbein J, Hortschansky P, Morton CO, Brakhage AA, Topp M, Einsele H, Wurster S, Loeffler J | 2021 | CcpA- and Shm2-pulsed myeloid dendritic cells induce T-cell activation and enhance the neutrophilic oxidative burst response to Aspergillus fumigatus. | Front Immunol 12: 659752 | PubMed |
| Brakhage AA, Zimmermann AK, Rivieccio F, Visser C, Blango MG | 2021 | Host-derived extracellular vesicles for antimicrobial defense. | microLife 2: uqab003 | microLife |
| Gupta SK, Srivastava M, Osmanoglu Ö, Xu Z, Brakhage AA, Dandekar T | 2021 | Aspergillus fumigatus versus Genus Aspergillus: Conservation, adaptive evolution, and specific virulence genes. | Microorganisms 9: 2014 | PubMed |
| Mogavero S, Sauer FM, Brunke S, Allert S, Schulz D, Wisgott S, Jablonowski N, Elshafee O, Krüger T, Kniemeyer O, Brakhage AA, Naglik JR, Dolk E, Hube B | 2021 | Candidalysin delivery to the invasion pocket is critical for host epithelial damage induced by Candida albicans. | Cell microbiol 23: e13378 | PubMed |
| Ewald J, Rivieccio F, Radosa L, Schuster S, Brakhage AA, Kaleta C | 2021 | Dynamic optimization reveals alveolar epithelial cells as key mediators of host defense in invasive aspergillosis. | PLoS Comp Bio 17: e1009645 | PubMed |
| Wich M, Greim S, Ferreira-Gomes M, Krüger T, Kniemeyer O, Brakhage AA, Jacobsen ID, Hube B, Jungnickel B | 2021 | Functionality of the human antibody response to Candida albicans. | Virulence 12: 3137-48 | PubMed |
| López-Berges MS, Scheven MT, Hortschansky P, Misslinger M, Baldin C, Gsaller F, Werner ER, Krüger T, Kniemeyer O, Weber J, Brakhage AA, Haas H | 2021 | The bZIP Transcription Factor HapX Is Post-Translationally Regulated to Control Iron Homeostasis in Aspergillus fumigatus. | Int J Mol Sci. 22(14): 7739 | PubMed |
| Radosa S, Sprague JL, Lau SH, Tóth R, Linde J, Krüger T, Sprenger M, Kasper L, Westermann M, Kniemeyer O, Hube B, Brakhage AA, Gácser A, Hillmann F | 2021 | The fungivorous amoeba Protostelium aurantium targets redox homeostasis and cell wall integrity during intracellular killing of Candida parapsilosis. | Cell Microbiol. 23(11): e13389 | PubMed |
| Shopova IA, Belyaev I, Dasari P, Jahreis S, Stroe MC, Cseresnyés Z, Zimmermann AK, Medyukhina A, Svensson CM, Krüger T, Szeifert V, Nietzsche S, Conrad T, Blango MG, Kniemeyer O, Lilienfeld-Toal Mv, Zipfel PF, Ligeti E, Figge MT, Brakhage AA | 2020 | Human neutrophils produce antifungal extracellular vesicles against Aspergillus fumigatus. | mBio 11: e00596-20 | PubMed |
| Blango MG, Pschibul A, Rivieccio F, Krüger T, Muhammad R, Jia L, Zheng T, Goldmann M, Voltersen V, Li Jun, Panagiotou G, Kniemeyer O, Brakhage AA | 2020 | Dynamic surface proteomes of allergenic fungal conidia. | J Proteome Res 19: 2092-104 | PubMed |
| Hassan MIA, Kruse J, Krüger T, Dahse HM, Cseresnyés Z, Blango MG, Slevogt H, Hörhold F, Ast V, König R, Figge MT, Kniemeyer O, Brakhage AA, Voigt K | 2020 | Functional surface proteomic profiling reveals the host heat-shock protein A8 as a mediator of Lichtheimia corymbifera recognition by murine alveolar macrophages. | Environ Microbiol 22: 3722-40 | PubMed |
| Ruben S, Garbe E, Mogavero S, Albrecht-Eckardt D, Hellwig D, Häder A, Krüger T, Gerth K, Jacobsen ID, Elshafee O, Brunke S, Hünniger K, Kniemeyer O, Brakhage AA, Morschhäuser J, Hube B, Vylkova S, Kurzai O, Martin R | 2020 | Ahr1 and Tup1 contribute to the transcriptional control of virulence-associated genes in Candida albicans. | mBio 11: e00206-20 | PubMed |
| Gonçalves SM, Duarte-Oliveira C, Campos CF, Aimanianda V, Ter Horst R, Leite L, Mercier T, Pereira P, Fernández-García M, Antunes D, Rodrigues CS, Barbosa-Matos C, Gaifem J, Mesquita I, Marques A, Osório NS, Torrado E, Rodrigues F, Costa S, Joosten LA, Lagrou K, Maertens J, Lacerda JF, Campos A Jr, Brown GD, Brakhage AA, Barbas C, Silvestre R, van de Veerdonk FL, Chamilos G, Netea MG, Latgé JP, Cunha C, Carvalho A | 2020 | Phagosomal removal of fungal melanin reprograms macrophage metabolism to promote antifungal immunity. | Nat Commun 11: 2282 | PubMed |
| Jia LJ, Krüger T, Blango MG, von Eggeling F, Kniemeyer O, Brakhage AA | 2020 | Biotinylated surfome profiling identifies potential biomarkers for diagnosis and therapy of Aspergillus fumigatus infection. | mSphere 5: e00535-20 | PubMed |
| Schmidt F, Thywißen A, Goldmann M, Cunha C, Cseresnyés Z, Schmidt H, Rafiq M, Galiani S, Gräler MH, Chamilos G, Lacerda JF, Campos Jr A, Eggeling C, Figge MT, Heinekamp T, Filler SG, Carvalho A, Brakhage AA | 2020 | Flotillin-dependent membrane microdomains are required for functional phagolysosomes against fungal infections. | Cell Rep 32: 108017 | PubMed |
| Dasari P, Koleci N, Shopova I, Wartenberg D, Beyersdorf N, Dietrich S, Sahagún-Ruiz A, Figge MT, Skerka C, Brakhage AA, Zipfel PF | 2019 | Enolase from Aspergillus fumigatus is a moonlighting and immune evasion protein that binds the human plasma complement protein factors H, FHL-1, C4BP, and Plasminogen. | Front Immunol 10: 2573 | PubMed |
| Satish S, Jimenez-Ortigosa C, Zhao Y, Lee MH, Dolgov E, Krüger T, Park S, Denning DW, Kniemeyer O, Brakhage AA, Perlin DS | 2019 | Stress-induced changes in the lipid microenvironment of β-(1,3)-D-glucan synthase cause clinically important echinocandin resistance in Aspergillus fumigatus. | mBio 10: e00779-19 | PubMed |
| König S, Pace S, Pein H, Heinekamp T, Kramer J, Romp E, Straßburger M, Troisi F, Proschak A, Dworschak J, Scherlach K, Rossi A, Sautebin L, Haeggström JZ, Hertweck C, Brakhage AA, Gerstmeier J, Proschak E, Werz O | 2019 | Gliotoxin from Aspergillus fumigatus abrogates leukotriene B4 formation through inhibition of leukotriene A4 hydrolase | Cell Chem Biol 26: 1-11 | PubMed |
| Bacher P, Hohnstein T, Beerbaum E, Röcker M, Kaufmann S, Brandt C, Röhmel J, Stervbo U, Nienen M, Babel N, Milleck J, Assenmacher M, Cornely OA, Heine G, Worm M, Creutz P, Tabeling C, Ruwwe-Glösenkamp C, Sander LE, Brunke S, Hube B, Blango M, Kniemeyer O, Brakhage AA, Schwarz C, Scheffold A | 2019 | Human anti-fungal Th17 immunity and pathology rely on cross-reactivity against Candida albicans. | Cell 176: 1340-55 | PubMed |
| Manfiolli AO, Siqueira FS, Dos Reis TF, Van Dijck P, Schrevens S, Hoefgen S, Föge M, Straßburger M, de Assis LJ, Heinekamp T, Rocha MC, Janevska S, Brakhage AA, Malavazi I, Goldman GH, Valiante V | 2019 | Mitogen-activated protein kinase cross-talk interaction modulates the production of melanins in Aspergillus fumigatus. | mBio 10: e00215-00219 | PubMed |
| Nossmann M, Boysen JM, Krüger T, König CC, Hillmann F, Munder T, Brakhage AA | 2019 | Yeast two-hybrid screening reveals a dual function for the histone acetyltransferase GcnE by controlling glutamine synthesis and development in Aspergillus fumigatus. | Curr Genet 65: 523-38 | PubMed |
| Shekhova E, Ivanova L, Krüger T, Stroe MC, Macheleidt J, Kniemeyer O, Brakhage AA | 2019 | Redox proteomic analysis reveals oxidative modifications of proteins by increased levels of intracellular reactive oxygen species during hypoxia adaptation of Aspergillus fumigatus. | Proteomics 19: 1800339 | PubMed |
| Schmidt H, Vlaic S, Krüger T, Schmidt F, Balkenhol J, Dandekar T, Guthke R, Kniemeyer O, Heinekamp T, Brakhage AA | 2018 | Proteomics of Aspergillus fumigatus conidia-containing phagolysosomes identifies processes governing immune evasion. | Mol Cell Proteomics 17: 1084-96 | PubMed |
| Dasari P, Shopova IA, Stroe M, Wartenberg D, Dahse HM, Beyersdorf N, Hortschansky P, Dietrich S, Cseresnyés Z, Figge MT, Westermann M, Skerka C, Brakhage AA, Zipfel PF | 2018 | Aspf2 from Aspergillus fumigatus recruits human immune regulators for immune evasion and cell damage. | Front Immunol 9: 1635 | Frontiers |
| Conrad T, Kniemeyer O, Henkel SG, Krüger T, Mattern DJ, Valiante V, Guthke R, Jacobsen ID, Brakhage AA, Vlaic S, Linde J | 2018 | Module-detection approaches for the integration of multilevel omics data highlight the comprehensive response of Aspergillus fumigatus to caspofungin. | BMC Syst Biol 12: 88 | PubMed |
| Van Dijck P, Sjollema J, Cammue BP, Lagrou K, Berman J, d'Enfert C, Andes DR, Arendrup MC, Brakhage AA, Calderone R, Cantón E, Coenye T, Cos P, Cowen LE, Edgerton M, Espinel-Ingroff A, Filler SG, Ghannoum M, Gow NAR, Haas H, Jabra-Rizk MA, Johnson EM, Lockhart SR, Lopez-Ribot JL, Maertens J, Munro CA, Nett JE, Nobile CJ, Pfaller MA, Ramage G, Sanglard D, Sanguinetti M, Spriet I, Verweij PE, Warris A, Wauters J, Yeaman MR, Zaat SAJ, Thevissen K | 2018 | Methodologies for in vitro and in vivo evaluation of efficacy of antifungal and antibiofilm agents and surface coatings against fungal biofilms. | Microb Cell 5: 300-26 (Review) | PubMed |
| Noßmann M, Pieper J, Hillmann F, Brakhage AA, MunderT | 2018 | Generation of an arginine-tRNA-adapted Saccharomyces cerevisiae strain for effective heterologous protein expression. | Curr Genet 64: 589-98 | PubMed |
| Voltersen V, Blango MG, Herrmann S, Schmidt F, Heinekamp T, Strassburger M, Krüger T, Bacher P, Lother J, Weiss E, Hünniger K, Liu H, Hortschansky P, Scheffold A, Löffler J, Krappmann S, Nietzsche S, Kurzai O, Einsele H, Kniemeyer O, Filler SG, Reichard U, Brakhage AA | 2018 | Proteome analysis reveals the conidial surface protein CcpA essential for virulence of the pathogenic fungus Aspergillus fumigatus. | mBio 9: e01557-18 | PubMed |
| Bergfeld A, Dasari P, Werner S, Hughes TR, Song WC, Hortschansky P, Brakhage AA, Hünig T, Zipfel PF, Beyersdorf N | 2017 | Direct binding of the pH-regulated protein 1 (Pra1) from Candida albicans inhibits cytokine secretion by mouse CD4+ T cells. | Front Microbiol 8: 844 | PubMed |
| Pohlers S, Martin R, Krüger T, Hellwig D, Hänel F, Kniemeyer O, Saluz HP, Van Dijck P, Ernst JF, Brakhage A, Mühlschlegel FA, Kurzai O | 2017 | Lipid signaling via Pkh1/2 regulates fungal CO2 sensing through the kinase Sch9. | mBio 8: e02211-16 | PubMed |
| Johns A, Scharf DH, Gsaller F, Schmidt H, Heinekamp T, Straßburger M, Oliver JD, Birch M, Beckmann N, Dobb KS, Gilsenan J, Rash B, Bignell E, Brakhage AA, Bromley MJ | 2017 | A nonredundant Phosphopantetheinyl transferase, PptA, is a novel antifungal target that directs secondary metabolite, Siderophore, and Lysine biosynthesis in Aspergillus fumigatus and is critical for pathogenicity. | mBio 8: e01504-16 | PubMed |
| de Vries RP, Riley R, Wiebenga A, Aguilar-Osorio G, Amillis S, Uchima CA, Anderluh G, Asadollahi M, Askin M, Barry K, Battaglia E, Bayram Ö, Benocci T, Braus-Stromeyer SA, Caldana C, Cánovas D, Cerqueira GC, Chen F, Chen W, Choi C, Clum A, Dos Santos RA, Damásio AR, Diallinas G, Emri T, Fekete E, Flipphi M, Freyberg S, Gallo A, Gournas C, Habgood R, Hainaut M, Harispe ML, Henrissat B, Hildén KS, Hope R, Hossain A, Karabika E, Karaffa L, Karányi Z, Kraševec N, Kuo A, Kusch H, LaButti K, Lagendijk EL, Lapidus A, Levasseur A, Lindquist E, Lipzen A, Logrieco AF, MacCabe A, Mäkelä MR, Malavazi I, Melin P, Meyer V, Mielnichuk N, Miskei M, Molnár ÁP, Mulé G, Ngan CY, Orejas M, Orosz E, Ouedraogo JP, Overkamp KM, Park HS, Perrone G, Piumi F, Punt PJ, Ram AF, Ramón A, Rauscher S, Record E, Riaño-Pachón DM, Robert V, Röhrig J, Ruller R, Salamov A, Salih NS, Samson RA, Sándor E, Sanguinetti M, Schütze T, Sepčić K, Shelest E, Sherlock G, Sophianopoulou V, Squina FM, Sun H, Susca A, Todd RB, Tsang A, Unkles SE, van de Wiele N, van Rossen-Uffink D, Oliveira JV, Vesth TC, Visser J, Yu JH, Zhou M, Andersen MR, Archer DB, Baker SE, Benoit I, Brakhage AA, Braus GH, Fischer R, Frisvad JC, Goldman GH, Houbraken J, Oakley B, Pócsi I, Scazzocchio C, Seiboth B, vanKuyk PA, Wortman J, Dyer PS, Grigoriev IV | 2017 | Comparative genomics reveals high biological diversity and specific adaptations in the industrially and medically important fungal genus Aspergillus. | Genome Biol 18: 28 | PubMed |
| Kroll K, Shekhova E, Mattern DJ, Thywissen A, Jacobsen ID, Strassburger M, Heinekamp T, Shelest E, Brakhage AA, Kniemeyer O | 2016 | The hypoxia-induced dehydrogenase HorA is required for coenzyme Q10 biosynthesis, azole sensitivity and virulence of Aspergillus fumigatus. | Mol Microbiol 101: 92-108 | PubMed |
| Teutschbein J, Simon S, Lother J, Springer J, Hortschansky P, Morton CO, Löffler J, Einsele H, Conneally E, Rogers TR, Guthke R, Brakhage AA, Kniemeyer O | 2016 | Proteomic profiling of serological responses to Aspergillus fumigatus antigens in patients with invasive aspergillosis. | J Proteome Res 15: 1580-91 | PubMed |
| Horn F, Linde J, Mattern DJ, Walther G, Guthke R, Scherlach K, Martin K, Brakhage AA, Petzke L, Valiante V | 2016 | Draft genome sequences of fungus Aspergillus calidoustus. | Genome Announc 4: e00102-16 | PubMed |
| Bacher P, Heinrich F, Stervbo U, Nienen M, Vahldieck M, Iwert C, Vogt K, Kollet J, Babel N, Sawitzki B, Schwarz C, Bereswill S, Heimesaat MM, Heine G, Gadermaier G, Asam C, Assenmacher M, Kniemeyer O, Brakhage AA, Ferreira F, Wallner M, Worm M, Scheffold A | 2016 | Regulatory T cell specificity directs tolerance versus allergy against aeroantigens in humans. | Cell 167: 1067-78.e16 | PubMed |
| Valiante V, Baldin C, Hortschansky P, Jain R, Thywißen A, Straßburger M, Heinekamp T, Brakhage AA | 2016 | The Aspergillus fumigatus conidial melanin production is regulated by the bifunctional bHLH DevR and MADS-box RlmA transcription factors. | Mol Microbiol 102: 321-335. | PubMed |
| Pollmächer J, Timme S, Schuster S, Brakhage AA, Zipfel PF, Figge MT | 2016 | Deciphering the counterplay of Aspergillus fumigatus infection and host inflammation by evolutionary games on graphs. | Sci Rep 6: 27807 | PubMed |
| Luo T, Krüger T, Knüpfer U, Kasper L, Wielsch N, Hube B, Kortgen A, Bauer M, Giamarellos-Bourboulis EJ, Dimopoulos G, Brakhage AA, Kniemeyer O | 2016 | Immunoproteomic analysis of antibody responses to extracellular proteins of Candida albicans revealing the importance of glycosylation for antigen recognition. | J Proteome Res 15: 2394-406 | PubMed |
| Kniemeyer O, Ebel F, Krüger T, Bacher P, Scheffold A, Luo T, Strassburger M, Brakhage AA | 2016 | Immunoproteomics of Aspergillus for the development of biomarkers and immunotherapies. | Proteomics Clin Appl 10: 910-21 | PubMed |
| Horn F, Habel A, Scharf DH, Dworschak J, Brakhage AA, Guthke R, Hertweck C, Linde J | 2015 | Draft genome sequence and gene annotation of the entomopathogenic fungus Verticillium hemipterigenum. | Genome Announc 3: e01439-14 | PubMed |
| Baldin C, Valiante V, Krüger T, Schafferer L, Haas H, Kniemeyer O, Brakhage AA | 2015 | Comparative proteomics of a tor inducible Aspergillus fumigatus mutant reveals involvement of the tor kinase in iron regulation. | Proteomics 15: 2230-43 | PubMed |
| Kraibooj K, Schoeler H, Svensson CM, Brakhage AA, Figge MT | 2015 | Automated quantification of the phagocytosis of Aspergillus fumigatus conidia by a novel image analysis algorithm. | Front Microbiol 6: 549 | PubMed |
| Altwasser R, Baldin C, Weber J, Guthke R, Kniemeyer O, Brakhage AA, Linde J, Valiante V | 2015 | Network modeling reveals cross talk of MAP kinases during adaptation to caspofungin stress in Aspergillus fumigatus. | PLoS One 10: e0136932 | PubMed |
| Horn F, Linde J, Mattern DJ, Walther G, Guthke R, Brakhage AA, Valiante V | 2015 | Draft genome sequence of the fungus Penicillium brasilianum MG11. | Genome Announc 3: e00724-15 | PubMed |
| Lin CJ, Sasse C, Valerius O, Irmer H, Heinekamp T, Straßburger M, Tran VT, Herzog B, Braus-Stromeyer SA, Braus GH | 2015 | Transcription factor SomA Is Required for Adhesion, Development and Virulence of the Human Pathogen Aspergillus fumigatus. | PLoS Pathog 11: e1005205 | PubMed |
| Mattern DJ, Schoeler H, Weber J, Novohradská S, Kraibooj K, Dahse HM, Hillmann F, Valiante V, Figge MT, Brakhage AA | 2015 | Identification of the antiphagocytic trypacidin gene cluster in the human-pathogenic fungus Aspergillus fumigatus. | Appl Microbiol Biotechnol 99: 10151-61 | PubMed |
| Krüger T, Luo T, Schmidt H, Shopova I, Kniemeyer O | 2015 | Challenges and strategies for proteome analysis of the interaction of human pathogenic fungi with host immune cells. | Proteomes 3: 467-95 | PubMed |
| Heinekamp T, Schmidt H, Lapp K, Pähtz V, Shopova I, Köster-Eiserfunke N, Krüger T, Kniemeyer O, Brakhage AA | 2015 | Interference of Aspergillus fumigatus with the immune response. | Semin Immunopathol 37: 141-52 | PubMed |
| Hillmann F, Linde J, Beckmann N, Cyrulies M, Strassburger M, Heinekamp T, Haas H, Guthke R, Kniemeyer O, Brakhage AA | 2014 | The novel globin protein fungoglobin is involved in low oxygen adaptation of Aspergillus fumigatus. | Mol Microbiol 93: 539-53 | PubMed |
| Horn F, Schroeckh V, Netzker T, Guthke R, Brakhage AA, Linde J | 2014 | Draft genome sequence of Streptomyces iranensis. | Genome Announc 2: e00616-14 | PubMed |
| Lapp K, Vödisch M, Kroll K, Strassburger M, Kniemeyer O, Heinekamp T, Brakhage AA | 2014 | Characterisation of the Aspergillus fumigatus detoxification systems for reactive nitrogen intermediates and impact on virulence. | Front Microbiol 5: 469 | PubMed |
| Scharf DH, Heinekamp T, Brakhage AA | 2014 | Human and plant fungal pathogens: the role of secondary metabolites. | PLoS Pathog 10: e1003859 | PLoS Pathog |
| Schwartze VU, Santiago AL, Jacobsen ID, Voigt K | 2014 | The pathogenic potential of the Lichtheimia genus revisited: Lichtheimia brasiliensis is a novel, non-pathogenic species. | Mycoses 57 Suppl 3: 128-31 | PubMed |

