Project B3
Predictive modeling of host-fungal pathogen interactions by reconstruction of gene regulatory networks
This project was completed after the first funding period.
The objective of the project B3 was to investigate the interaction of the human pathogenic fungi Candida albicans and Aspergillus fumigatus with human immune cells by network modeling. Utilizing omics-data, we predicted gene regulatory networks by mathematical modeling. This was done in both genome-wide large-scale and small-scale networks, where the latter focused on a pre-defined question for the human-pathogenic fungi. The predictive network modeling of host-fungal pathogen interactions was based on both prior knowledge and high-throughput data. We employed comprehensive and integrative analysis and mathematical modeling of the transcriptome (RNA-Seq) and proteome of both, pathogen and host cells. We inferred gene regulatory network models using top down approaches which is hypothesis-free at the beginning and a prerequisite to avoid missing any components or trends by initial filtering and/or biased analysis of large-scale data sets available from the partners of the CRC/TR. The analyzed experimental data such as transcriptome and other genome-wide data as well as microbial, biochemical and clinical data, were generated by the members of the CRC/TR FungiNet and also retrieved from public databases. Prior knowledge was semi-automatically extracted from databases and literature repositories.
The resulting gene regulatory networks were used for optimal design of follow-up experiments and to support the development of markers for improved diagnosis of fungal infections, in particular invasive aspergillosis. Furthermore, non-coding RNAs (ncRNAs) were investigated in order to determine functions of ncRNAs in fungus-host interaction networks. In collaboration with partners from the CRC/TR FungiNet, we conducted an expression analysis of human miRNAs of dendritic cells, comparing infections of A. fumigatus and C. albicans with lipopolysaccharide (LPS) treatment to simulate bacterial infections. By performing small RNA-sequencing, we identified miRNAs that are differentially expressed during fungal infections. We determined putative target mRNAs of these miRNAs based on prediction tools and a database for validated miRNA-target-interactions.
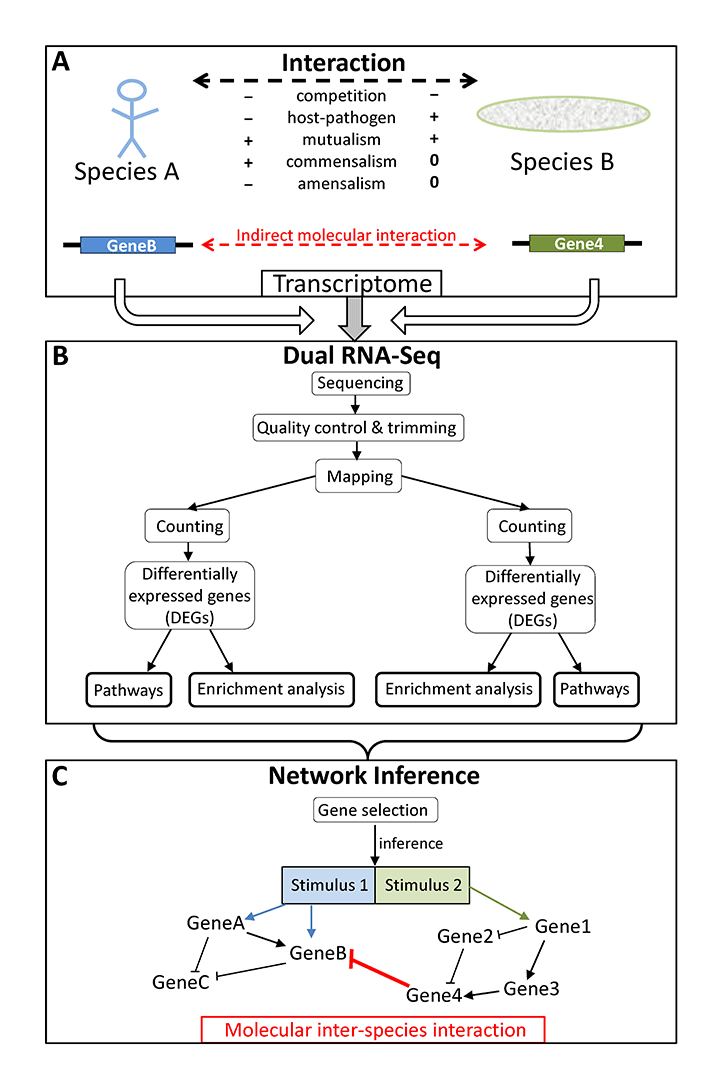
Schematic overview of (A) possible interactions between organisms of two different species influencing each others transcriptome, (B) the processing of dual RNA-seq data, and (C) the result of network inference showing the indirect molecular interaction between two genes from two different species (red bar) and molecular intra-species interactions (black arrows and bars) within each of the two species (from Schulze et al. 2016).

Prof. Dr. Manja Marz
Faculty of Mathematics and Computer Science Bioinformatics/High Throughput Analysis
Friedrich Schiller University Jena
| Author | Year | Title | Journal | Links |
|---|---|---|---|---|
| Kämmer P, McNamara S, Wolf T, Conrad T, Allert S, Gerwien F, Hünniger K, Kurzai O, Guthke R, Hube B, Linde J, Brunke S | 2020 | Survival strategies of pathogenic Candida species in human blood show independent and specific adaptations. | mBio 11: e02435-20 | PubMed |
| Müller MM, Lehmann R, Klassert TE, Reifenstein S, Conrad T, Moore C, Kuhn A, Behnert A, Guthke R, Driesch D, Slevogt H | 2017 | Global analysis of glycoproteins identifies markers of endotoxin tolerant monocytes and GPR84 as a modulator of TNFα expression. | Sci Rep 7: 838 | PubMed |
| Klassert TE, Bräuer J, Hölzer M, Stock M, Riege K, Zubiría-Barrera C, Müller MM, Rummler S, Skerka C, Marz M, Slevogt H | 2017 | Differential effects of vitamins A and D on the transcriptional landscape of human monocytes during infection. | Sci Rep 7: 40599 | PubMed |
| Riege K, Hölzer M, Klassert TE, Barth E, Bräuer J, Collatz M, Hufsky F, Mostajo N, Stock M, Vogel B, Slevogt H, Marz M | 2017 | Massive effect on LncRNAs in human monocytes during fungal and bacterial infections and in response to vitamins A and D. | Sci Rep 7: 40598 | PubMed |
| Dix A, Czakai K, Leonhardt I, Schäferhoff K, Bonin M, Guthke R, Einsele H, Kurzai O, Löffler J, Linde J | 2017 | Specific and novel microRNAs are regulated as response to fungal infection in human dendritic cells. | Front Microbiol 8: 270 | PubMed |
| Prauße MT, Schäuble S, Guthke R, Schuster S | 2016 | Computing the various pathways of penicillin synthesis and their molar yields. | Biotechnol Bioeng 113: 173-81 | PubMed |
| Durmuş S, Çakır T, Guthke R | 2016 | Editorial: Computational systems biology of pathogen-host interactions. | Front Microbiol 7: 21 | PubMed |
| Böhringer M, Pohlers S, Schulze S, Albrecht-Eckardt D, Piegsa J, Weber M, Martin R, Hünniger K, Linde J, Guthke R, Kurzai O | 2016 | Candida albicans infection leads to barrier breakdown and a MAPK/NF-κB mediated stress response in the intestinal epithelial cell line C2BBe1. | Cell Microbiol 18: 889-904 | Cell Microbiol |
| Dix A, Czakai K, Springer J, Fliesser M, Bonin M, Guthke R, Schmitt AL, Einsele H, Linde J, Löffler J | 2016 | Genome-wide expression profiling reveals S100B as biomarker for invasive Aspergillosis. | Front Microbiol 7: 320 | PubMed |
| Kroll K, Shekhova E, Mattern DJ, Thywissen A, Jacobsen ID, Strassburger M, Heinekamp T, Shelest E, Brakhage AA, Kniemeyer O | 2016 | The hypoxia-induced dehydrogenase HorA is required for coenzyme Q10 biosynthesis, azole sensitivity and virulence of Aspergillus fumigatus. | Mol Microbiol 101: 92-108 | PubMed |
| Teutschbein J, Simon S, Lother J, Springer J, Hortschansky P, Morton CO, Löffler J, Einsele H, Conneally E, Rogers TR, Guthke R, Brakhage AA, Kniemeyer O | 2016 | Proteomic profiling of serological responses to Aspergillus fumigatus antigens in patients with invasive aspergillosis. | J Proteome Res 15: 1580-91 | PubMed |
| Dix A, Vlaic S, Guthke R, Linde J | 2016 | Use of systems biology to decipher host-microbial interaction networks and predict biomarkers. | Clin Microbiol Infect 22: 600-6 | PubMed |
| Guthke R, Gerber S, Conrad T, Vlaic S, Durmuş S, Çakır T, Sevilgen FE, Shelest E, Linde J | 2016 | Data-based reconstruction of gene regulatory networks of fungal pathogens. | Front Microbiol 7: 570 | PubMed |
| Schulze S, Schleicher J, Guthke R, Linde J | 2016 | How to predict molecular interactions between species? | Front Microbiol 7: 442 | PubMed |
| Czakai K, Leonhardt I, Dix A, Bonin M, Linde J, Einsele H, Kurzai O, Loeffler J | 2016 | Krüppel-like Factor 4 modulates interleukin-6 release in human dendritic cells after in vitro stimulation with Aspergillus fumigatus and Candida albicans | Sci Rep 6: 27990 | PubMed |
| Shelest E, Wingender E | 2016 | Editorial: Systems biology of transcription regulation. | Front Genet 7: 124 | PubMed |
| Schulze S, Henkel SG, Driesch D, Guthke R, Linde J | 2015 | Computational prediction of molecular pathogen-host interactions based on dual transcriptome data. | Front Microbiol 6: 65 | PubMed |
| Dix A, Hünniger K, Weber M, Guthke R, Kurzai O, Linde J | 2015 | Biomarker-based classification of bacterial and fungal whole-blood infections in a genome-wide expression study. | Front Microbiol 6: 171 | PubMed |
| Linde J, Schulze S, Henkel SG, Guthke R | 2015 | Data- and knowledge-based modeling of gene regulatory networks: An update. | EXCLI J 14: 346-78 | EXCLI Journal |
| Linde J, Duggan S, Weber M, Horn F, Sieber P, Hellwig D, Riege K, Marz M, Martin R, Guthke R, Kurzai O | 2015 | Defining the transcriptomic landscape of Candida glabrata by RNA-Seq. | Nucleic Acids Res 43: 1392-406 | PubMed |
| Durmuş S, Çakır T, Özgür A, Guthke R | 2015 | A review on computational systems biology of pathogen-host interactions. | Front Microbiol 6: 235 | PubMed |
| Ramachandra S, Linde J, Brock M, Guthke R, Hube B, Brunke S | 2014 | Regulatory networks controlling nitrogen sensing and uptake in Candida albicans. | PLoS One 9: e92734 | PubMed |